
Research Overview
Cellular protein concentrations are controlled through the rates of synthesis and degradation. Most regulated degradation in eukaryotic cells occurs via the Ubiquitin Proteasome System (UPS) which is responsible for regulating the concentration of thousands of cellular proteins as well as clearing damaged and misfolded proteins from the cell. At the center of the UPS is the 26S proteasome, a macromolecular proteolytic machine that maintains cellular homeostasis by recognizing and destroying proteins that have been targeted for degradation. To dissect molecular mechanisms of the UPS, we use experimental methods ranging from combinatorial protein engineering, quantitative biochemical assays, cell culture assays, genome-scale screens, and single molecule biophysics.
 |  |  |
|---|---|---|
 |  |  |
 |
Protein Targeting
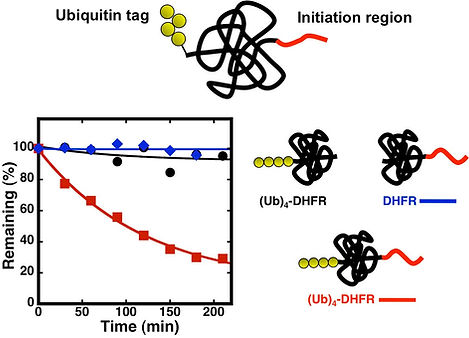
We found that ubiquitin tags by themselves are not sufficient to target folded proteins for proteasomal degradation (Prakash et al. Nat. Struct. Mol. Biol. 2004). Instead, degradation signals or degrons contain a second part in the form of an intrinsically disordered initiation regions in substrates that can serve as initiation sites for the proteasome. Thus, the proteasome binds to its substrates through the ubiquitin tag and engages them at the initiation site.
We investigate how the initiation site contributes to proteasome targeting. For example, we found that the initiation region has to be long enough (Fishbain et al. Nat. comm. 2011) and placed at the correct distance from the ubiquitin tag so that the proteasome can bind them both simultaneously (Inobe et al. Nat. Chem. Biol. 2011). Surprisingly, the proteasome also shows strong preferences for the amino acid sequence of the initiation region in the substrate. This raises the question of how the ubiquitin tag and the initiation region can compensate for each other. For example, can a substrate with a poor initiation region be degraded if it contains a better ubiquitin tag? Can a substrate with a very good initiation region be degraded even without a ubiquitin tag? Ultimately, we would like to understand the dynamics of how the two components interplay to contribute to efficient yet specific proteasomal degradation.
Partial degradation
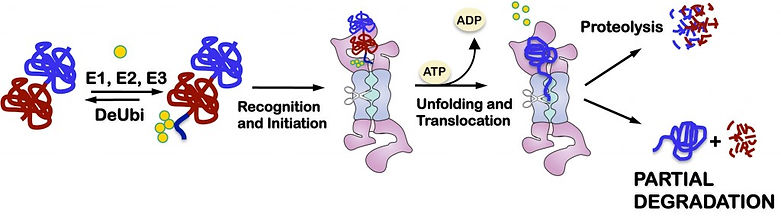
Once the proteasome has engaged a substrate it runs along the protein’s polypeptide chain degrading it sequentially and processively from the initiation site (Lee et al. Mol. Cell. 2001) into small peptides of some 10 amino acids in lengths. This prevents the formation of protein fragments with unwanted biological activities. However, there are a few examples where the proteasome stops degradation part way through a polypeptide chain and releases a partially degraded protein. We think that this partial degradation occurs when an internal ubiquitination site sends the proteasome to run into a stop signal encoded in the substrate (Tian et al. Nat. Struct. Mol. Biol. 2005; Schrader et al. J. Biol. Chem. 2011). We are now looking for more natural examples of partial degradation and are using biophysical techniques to understand the mechanism.
Tool Development
We found that the ubiquitin tag and initiation region can work together in trans so that a ubiquitinated receptor can target a binding partner containing only the initiation region for proteolysis by the proteasome (Prakash et al. Nat. Chem. Biol. 5, 29 (2009)). This is surprising since several regulatory processes in the cell rely on the proteasome to remodel protein complexes by extracting a specific subunit while leaving the rest of the complex intact. We predict that the subunit specificity of the proteasome depends on its selection of the correct initiation site. It is possible that trans-targeting is used physiologically, for example by viruses that want to be sure to deplete a cellular protein by targeting it to the proteasome with an adaptor protein that binds proteasome and substrate simultaneously. Finally, it may be possible to construct artificial adaptors to deplete toxic proteins, to interfere in a cellular process by depleting a regulatory protein, or to redirect metabolic flow by removing a key enzyme in a pathway after a branch point.
